HOW TO AND EXAMPLE
Functionality
There are few operations to be carried out before to reproduce a PRS in your cohort, specifically:
- Get .ped and .map files
- v1.07 link: PLINK v1.07
- v1.9/2.0 link: PLINK v1.9/2.0 beta
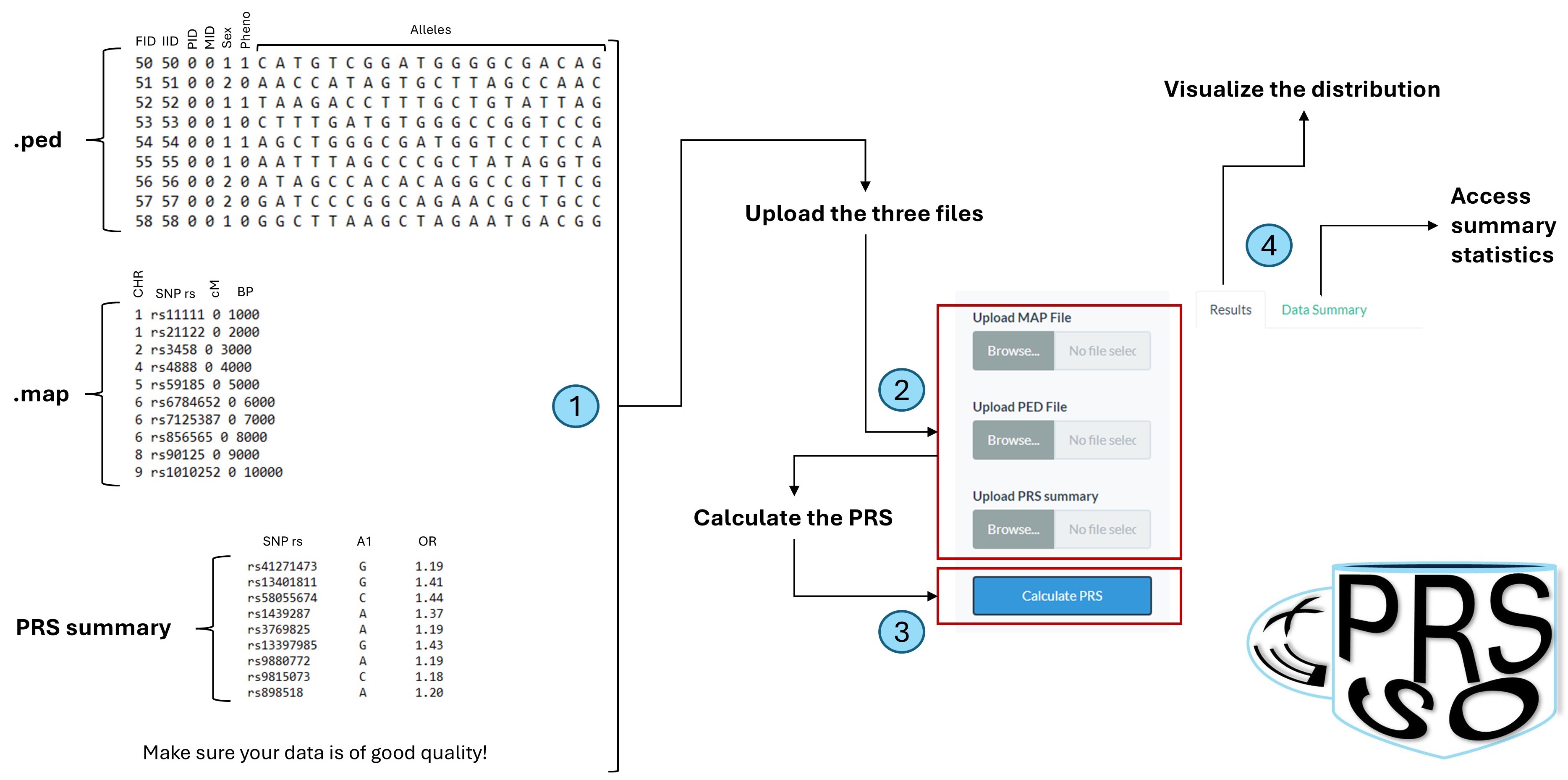
- Pedigree file: a text file reporting pedigree information and genotype calls. A pedigree file is a white-space (space or tab) delimited file with extension .ped, has no header, each row represents a single individual, and is composed by these columns:
- Family ID (FID)
- Individual ID (IID)
- Paternal ID (PID)
- Maternal ID (MID)
- Sex
- Phenotype
- Alleles (variable number of columns)
- Map file: is a text file that must contain 4 columns:
- Chromosome: a number from 1 to 22, X or Y
- SNP reference sequence: such as rsXXXXX
- Genetic distance
- BP position
- Download the PRS summary
- Upload your data
- Compute the score
These file can be computed using PLINK, the free open-source whole genome association analysis toolset.
Here's a brief description of the required structure for the .ped and .map files.
Before to upload .ped and .map files in exPRSso it is advisable to perform some quality controls (QCs) on your data. An example of data QCs is given here.
The PRS summary can be gathered by different sources, such as PGScatalog. This is an online repository where PRSs for a wide range of traits can be found. Additional information are reported on their page.
It is important that the PRS summary is uploaded as a text file without a header and with these columns: SNP, A1 and OR. Always check your PRS summary before to upload them in exPRSso.
Once you have collected the three required files, you can upload them in exPRSso. To do that, just go to the "exPRSso" section at the top of this page. You will be redirected to the application page. Here, you will find three upload buttons, one for each file to be uploaded.
Click on the "Calculate PRS" button and wait for the results to be ready!
Workflow